
Song Hong, Ph.D.
Song Hong, Ph.D.
Professor of Research, Neuroscience, Biochemistry, Cell and Molecular Biology, and Ophthalmology
Professor of Research, Neuroscience, Biochemistry, Cell and Molecular Biology, and Ophthalmology
Neuroscience Center of Excellence
School of Medicine, LSU Health
8th floor, Room 8A7
2020 Gravier St., Suite D
New Orleans, LA 70112
Phone: (504) 599-0335 (O);
shong@lsuhsc.edu
Degrees
1996-2001: Postdoctoral training - Cornell University,
New York
1996: PhD - The University of Georgia
Bio
Positions
2023-Present: Professor, Neuroscience, Biochemistry, Cell & Molecular Biology, and Ophthalmology.
Neuroscience Center of Excellence and Dept of Ophthamology
LSU Health Sciences Center, New Orleans, LA
2012-2023: Associate Professor, Neuroscience, Biochemistry, Cell & Molecular Biology, and Ophthalmology.
Neuroscience Center of Excellence and Dept of Ophthamology
LSU Health Sciences Center, New Orleans, LA
2018-present: Adjunct faculty, Diabetes Research, Tulane Medical School, Tulane University
2006-2012: Assistant Professor of Neuroscience, Biochemistry, Cell and Molecular Biology, and Ophthalmology.
Neuroscience Center of Excellence and Dept of Ophthalmology
LSU Health Sciences Center, New Orleans, LA
2004-2006: Instructor (a junior faculty rank), Harvard Medical School, Boston, MA
2001-2004: Research fellow, Harvard Medical School, Boston, MA
Clinical Interests
Current Research:
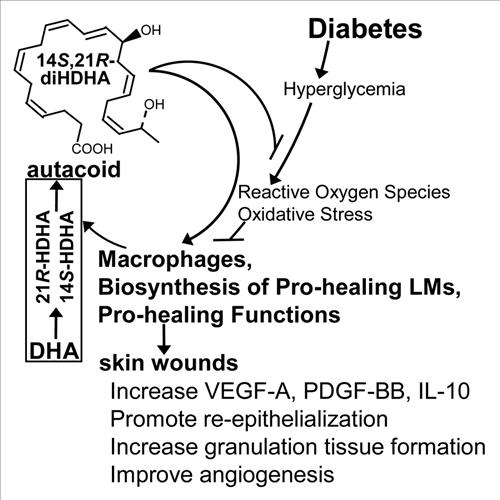
1) Our major objective is to elucidate new mechanisms and explore the therapeutic potential of endogenous reparative linflammation-resolving lipid mediators in diabetes and aging-impaired wound healing and in the regenerative functions of cells, angiogenesis, severe burns, and Alzheimer’s diseases. Our studies include the biochemical metabolic pathways and signaling of lipid mediators in diabetic wounds and burn injury. My work also includes the development of therapeutic leads and biomarkers for these disorders. We intend to decipher the roles of novel lipid mediators in the key cellular and molecular processes including inflammation, neurodegeneration, neurogenesis, innervation, phagocytosis/efferocytosis, microgliosis, astrogliosis, blood brainbarrier, vascularization/angiogenesis, epithelialization, and fibrosis. 2) We are also developing novel hydrogels, nanoparticles, and microparticles for wound healing and nerve regeneration. 3) We have been studying specialized pro-resolving and neuroprotective lipid mediators in Alzheimer's disease.
Research Interests
Keywords:
Wound healing, chronic wounds impaired by diabetes and aging, diabetic neurophathy, nerve regeneration and degeneration, Alzheimer's disease, neuron outgrowth, resolution of chronic inflammation, pro-healing lipid autacoids, pro-resolving reparative lipid mediators, mediator-lipidomic pathways, vasculogenesis, angiogenesis, fibrosis, phagocytosis, efferocytosis, wound healing, wound breaking strength, scarring, diabetic nephropathy, renal acute injury, leukocytes, neurons, microglial cells, astrocytes, spinal cord, dorsal root ganglions, macrophages, monocytes, dendritic cells, neutrophils, mesenchymal stem cells, adipose stem cells, platelets, endothelial cells, epithelial cells, T-cells, hydrogels, nanoparticles, and microparticles.
Research Interests and Goals:
Our long term goals are: 1) To delineate the actions, roles, and mechanisms of lipid mediators and mediator-lipidomic pathways that regulate wound healing and other reparative processes impaired by diabetes and aging as well as the pathogenesis of Alzheimer's disease. 2) To study the machanisms of these lipid mediators in other diabetic complications (neuropathy, nephropathy, and retinopathy) and Alzheimer's disease. 3) To provide new targets for better treatment of diabetic complications and Alzheimer's disease.
Our short-term goals are: 1) To elucidate the mechanism of action of endogenous reparative pro-reoslving lipid mediators in curbing the pathogesis of Alzheimer's disease and diabetic impairments of wound healing, in promoting re-innervation, vascularization, and integraty of blood brain barrier. We will also determine lipid-mediator based signatures of human non-healing chronic wounds. 2) To study the interaction of lipid mediators and stem cells or leukocytes in chronic wounds. 3) To study lipid mediators in ocular diseases.
Teaching Activities
-
NEURO 203 - Investigavtive Neuroscience
-
NCE 290 - Special Topics
- NEURO 250 - Molecular Neurobiology
-
CSI 100 and CSI 200 – School of Medicine House Mentorship Program
Committees & Administrative Responsibilities
- LSUHSC Research Advisory Committee
- LSUHSC Sabbatical Committee
- LSUHSC Graduate Advisory Committee
- LSUHSC Communication Committee
-
Current Editorial Board Membership
- International Journal of Stem cell research & Therapy
- Prostaglandins, Leukotrienes, Essential Fatty Acids
- International Journal of Surgery Research & Practice
- International Journal of Hematology Research
- SM Journal of Orthopedics
- Annals of Diabetes, Metabolic Disorders & Control
- The Open Biomarkers Journal
- Current Regenerative Medicine (CRM)
- Bioengineering
Selected Publications
Key Peer-reviewed Publications
(Selected from peer-reviewed publications)
- Baravkar SB, Lu Y, Zhao Q, Peng H, Zhou W, Hong S*. Rationally Designed Pentapeptide Analogs of Abeta19-23 Fragment as Potent Inhibitors of Abeta42 Aggregation. Molecules. 2025;30(9). Epub 20250507. doi: 10.3390/molecules30092071. PubMed PMID: 40363876; PMCID: PMC12073614.
- Hong S*, Baravkar SB, Lu Y, Masoud AR, Zhao Q, Zhou W. Molecular Modification of Queen Bee Acid and 10-Hydroxydecanoic Acid with Specific Tripeptides: Rational Design, Organic Synthesis, and Assessment for Prohealing and Antimicrobial Hydrogel Properties. Molecules. 2025;30(3). doi: 10.3390/molecules30030615. PubMed PMID: 39942719; PubMed Central PMCID: PMC 11819776.
- Shrivastava P, Lu Y, Su S, Kobayashi Y, Zhao Y, Lien N, Masoud AR, Lukiw WJ, Hong S*. Maresin-like 1 Ameliorates Neuropathology of Alzheimer's Disease in Brains of a Transgenic Mouse Model. Biomedicines. 2024;12(12):2865. doi: 10.3390/biomedicines12122865. PubMed PMID: 39767773; PubMed Central PMCID: PMC11673747.
- Lu, Y.; Su, S.; Chu, C.-C.; Kobayashi, Y.; Masoud, A.-R.; Peng, H.; Lien, N.; He, M.; Vuong, C.; Tran, R.; Hong, S.*, Amino Acid-Based Protein-Mimic Hydrogel Incorporating Pro-Regenerative Lipid Mediator and Microvascular Fragments Promotes the Healing of Deep Burn Wounds. Int. J. Mol. Sci. 2024, 25(19), 10378; DOI: https://doi.org/10.3390/ijms251910378
- Lu, Y.; Tian, H.; Peng, H.; Bazan, N.G.; Hong, S.*. Novel lipid mediator 7S,14R-docosahexaenoic acid: biogenesis and harnessing mesenchymal stem cells to ameliorate diabetic mellitus and retinal pericyte loss. Front Cell Dev Biol. 2024; 12: 1380059. PMCID: PMC10963555; PMID: 38533089
- Thamizhchelvan, A.M.; Masoud, A.-R.; Su, S.; Peng, H.; Kobayashi,Y.; Wang, Y.; Archer, N.; Hong, S.*. Bactericidal Efficacy of the Combination of Maresin-like Proresolving Mediators and Carbenicillin Action on Biofilm-Forming Burn Trauma Infection-Related Bacteria. International Journal of Molecular Sciences. 2024;25(5):2792. PMCID: PMC10932429; PMID:38474038
- Baravkar, S.B.; Lu, Y.; Masoud, A. R.; Zhao, Q.;, He, J.; Hong, S.*, Development of a Novel Covalently Bonded Conjugate of Caprylic Acid Tripeptide (Isoleucine–Leucine–Aspartic Acid) for Wound-Compatible and Injectable Hydrogel to Accelerate Healing. biomolecules. 2024, 14(1), 94. doi: 10.3390/biom14010094. PMCID: PMC10813153; PMID: 38254694.
- Hong, S.; Nagayach, A., Lu Y.; Peng, H.; Duong, Q.A.; Pham, N.B.; Vuong, C.A.; Bazan, N.G., A high fat, sugar, and salt Western diet induces motor-muscular and sensory dysfunctions and neurodegeneration in mice during aging: Ameliorative action of metformin. CNS Neurosci Ther. 2021 Dec;27(12):1458-1471. doi: 10.1111/cns.13726. PMCID: PMC8611779; PMID: 34510763.
- Ogawa, N.; Sone, S.; Hong, S.; Lu, Y.; Kobayashi, Y., Synthesis of Two Stereoisomers of Potentially Bioactive 13,19,20-Trihydroxy Derivative of Docosahexaenoic Acid. Synlett. 2020 Oct;31(17):1735-1739. PMCID: PMC8752060; PMID: 35023886.
- Hong, S.*, Lu, Y., Morita, M., Saito, S., Kobayashi, Y., Jun, B., Bazan, N.G., Xu, X., Wang, Y., Stereoselective Synthesis of Maresin-Like Lipid Mediators. Synlett. 2019, 30 (03) 343-347 PMCID: PMC6510270; PMID: 31086432.
- Alapure, B.V., Lu, Y., He, M., Chu, C.C., Peng, H., Muhale, F., Brewerton, Y.L., Bunnell, B., Hong, S.*, Accelerate Healing of Severe Burn Wounds by Mouse Bone Marrow Mesenchymal Stem Cell-Seeded Biodegradable Hydrogel Scaffold Synthesized from Arginine-Based Poly(ester amide) and Chitosan. Stem cells and development. 2018,27(23):1605-1620. PMCID:PMC6276600; PMID: 30215325.
- Alapure, B. V.; Lu, Y.; Peng, H.; Hong, S.*, Surgical denervation of specific cutaneous nerves impedes excisional wound healing of small animal ear pinnae. Mol Neurobiol, 2018, 55(2): 1236-1243. PMCID: PMC5577384; PMID: 28110472.
- Nishimura, K.; Sakaguchi, T.; Nanba, Y.; Suganuma, Y.; Morita, M.; Hong, S.; Lu, Y.; Jun, B.; Bazan, N. G.; Arita, M.; Kobayashi, Y., Stereoselective Total Synthesis of Macrophage-Produced Prohealing 14,21-Dihydroxy Docosahexaenoic Acids. The Journal of organic chemistry. 2018; 83(1):154-166. PMCID: PMC8607632; PMID: 29224348.
- Bazan, H. A.; Lu, Y.; Jun, B.; Fang, Z.; Woods, T. C.; Hong, S.*, Circulating inflammation-resolving lipid mediators RvD1 and DHA are decreased in patients with acutely symptomatic carotid disease. Prostaglandins, leukotrienes, and essential fatty acids. 2017; 125:43-47. PMCID: PMC5909403; PMID: 28987721.
- Hong, S. *; Tian, H.; Lu, Y.; Laborde, J. M.; Muhale, F. A.; Wang, Q.; Alapure, B. V.; Serhan, C. N.; Bazan, N. G., Neuroprotectin/Protectin D1: Endogenous Biosynthesis and Actions on Diabetic Macrophages in Promoting Wound Healing and Innervation Impaired by Diabetes. Am J Physiol Cell Physiol, 2014, 307, (11): C1058-67. PMCID: PMC4254953. PMID: 25273880.
- Hong, S.*; Lu, Y.; Tian, H.; Alapure, B. V.; Wang, Q.; Bonnell, B.; Laborde, J. M., Leukocytes and Platelets Produce Novel Maresin-like Docosanoids Structure, Biosynthesis, and Effect on Diabetes-impaired Macrophage Reparative Function. Chemistry & Biology (Cell at Cell Press), 2014, 21, (10): 1318-29. PMCID: PMC4224612. PMID:25200603.
- Hong, S.*; Alapure, B. V.; Lu, Y.; Tian, H.; Wang, Q., 12/15-Lipoxygenase deficiency reduces densities of mesenchymal stem cells in the dermis of wounded and unwounded skin. Br J Dermatol, 2014, 171, (1), 30-8. PMCID:PMC4114990. PMID: 24593251.
- Hong, S*.; Alapure, B. V.; Lu, Y.; Tian, H.; Wang, Q., Immunohistological localization of endogenous unlabeled stem cells in wounded skin. J Histochem Cytochem, 2014, 62, (4), 276-85. PMCID: PMC3966289. PMID: 24399040.
- Hong, S.*; Lu, Y., Omega-3 fatty acid-derived resolvins and protectins in inflammation resolution and leukocyte functions: targeting novel lipid mediator pathways in mitigation of acute kidney injury. Front Immunol,2013, 4, 13. PMCID:PMC3558681. PMID: 23386851.
- Tian, H.; Lu, Y.; Shah, S. P.; Wang, Q.; Hong, S.*, 14S,21R-dihydroxy-docosahexaenoic Acid Treatment Enhances Mesenchymal Stem Cell Amelioration of Renal Ischemia/Reperfusion Injury. Stem Cells Dev, 2012, 21, (7), 1187-99. PMCID:PMC3328765. PMID: 21846180.
- Bazan, N. G.; Eady, T. N.; Khoutorova, L.; Atkins, K. D.; Hong, S.; Lu, Y.; Zhang, C.; Jun, B.; Obenaus, A.; Fredman, G.; Zhu, M.; Winkler, J. W.; Petasis, N. A.; Serhan, C. N.; Belayev, L., Novel aspirin-triggered neuroprotectin D1 attenuates cerebral ischemic injury after experimental stroke. Exp Neurol,2012, 236, (1), 122-30. PMCID: PMC3409566. PMID: 22542947.
- Belayev, L.; Khoutorova, L.; Atkins, K. D.; Eady, T. N.; Hong, S.; Lu, Y.; Obenaus, A.; Bazan, N. G., Docosahexaenoic Acid Therapy of Experimental Ischemic Stroke. Transl Stroke Res,2011, 2(1), 33-41. PMCID: PMC3037476. PMID: 21423332.
- Tian, H.; Lu, Y.; Shah, S. P.; Hong, S.*, Autacoid 14S,21R-dihydroxy-docosahexaenoic acid counteracts diabetic impairment of macrophage prohealing functions. Am J Pathol, 2011, 179, (4), 1780-91.PMCID:PMC3181353. PMID: 21839062.
- Tian, H.; Lu, Y.; Shah, S. P.; Hong, S.*, 14S,21R-Dihydroxydocosahexaenoic Acid Remedies Impaired Healing and Mesenchymal Stem Cell Functions in Diabetic Wounds. J Biol Chem, 2011, 286, (6), 4443-53. PMCID:PMC3039401. PMID: 21112969.
- Tian, H.; Lu, Y.; Shah, S. P.; Hong, S.*, Novel 14S,21-dihydroxy-docosahexaenoic acid rescues wound healing and associated angiogenesis impaired by acute ethanol intoxication/exposure. J Cell Biochem, 2010, 111, (2), 266-73. PMCID:PMC3308707. PMID: 20506249.
- Lu, Y.; Tian, H.; Hong, S.*, Novel 14,21-dihydroxy-docosahexaenoic acids: structures, formation pathways, and enhancement of wound healing. J Lipid Res2010, 51, (5), 923-32. PMCID:PMC2853460. PMID:19965612.
- Marcheselli, V. L.; Mukherjee, P. K.; Arita, M.; Hong, S.; Antony, R.; Sheets, K.; Winkler, J. W.; Petasis, N. A.; Serhan, C. N.; Bazan, N. G., Neuroprotectin D1/protectin D1 stereoselective and specific binding with human retinal pigment epithelial cells and neutrophils. Prostaglandins Leukot Essent Fatty Acids, 2010, 82, (1), 27-34. PMCID:PMC2822836.PMID: 19931440,
- Tian, H.; Lu, Y.; Sherwood, A. M.; Hongqian, D.; Hong, S.*, Resolvins E1 and D1 in choroid-retinal endothelial cells and leukocytes: biosynthesis and mechanisms of anti-inflammatory actions. Invest Ophthalmol Vis Sci, 2009, 50, (8), 3613-20. PMID:19443724.
- Bazan, H. A.; Lu, Y.; Thoppil, D.; Fitzgerald, T. N.; Hong, S.; Dardik, A., Diminished omega-3 fatty acids are associated with carotid plaques from neurologically symptomatic patients: implications for carotid interventions. Vascul Pharmacol, 2009, 51, (5-6), 331-336. PMCID: PMC2786054. PMID: 19733689.
- Hong, S.; Porter, T. F.; Lu, Y.; Oh, S. F.; Pillai, P. S.; Serhan, C. N., Resolvin E1 metabolome in local inactivation during inflammation-resolution. J Immunol, 2008, 180, (5), 3512-9. PMID:18292578.
- Bazan, N. G.; Marcheselli, V. L.; Lu, Y.; Hong, S.; Jackson, F., Lipidomic Approaches to Neuroprotection Signaling in the Retinal Pigment Epithelium. In Signal Transduction in the Retina, Fliesler, S. J.; Kisselev, O. G., Eds. CRC Press: 2007; pp 345-374.
- Hong, S.; Lu, Y.; Yang, R.; Gotlinger, K. H.; Petasis, N. A.; Serhan, C. N., Resolvin D1, protectin D1, and related docosahexaenoic acid-derived products: Analysis via electrospray/low energy tandem mass spectrometry based on spectra and fragmentation mechanisms. J Am Soc Mass Spectrom, 2007, 18, (1), 128-44. PMCID:PMC2763184. PMID:17055291.
- Lu, Y.*; Hong, S.*; Yang, R.; Uddin, J.; Gotlinger, K. H.; Petasis, N. A.; Serhan, C. N., Identification of endogenous resolvin E1 and other lipid mediators derived from eicosapentaenoic acid via electrospray low-energy tandem mass spectrometry: spectra and fragmentation mechanisms. Rapid Commun Mass Spectrom, 2007, 21, (1), 7-22. PMID:17131464.(*Share 1st authorship).
- Connor, K. M.; SanGiovanni, J. P.; Lofqvist, C.; Aderman, C. M.; Chen, J.; Higuchi, A.; Hong, S.; Pravda, E. A.; Majchrzak, S.; Carper, D.; Hellstrom, A.; Kang, J. X.; Chew, E. Y.; Salem, N., Jr.; Serhan, C. N.; Smith, L. E., Increased dietary intake of omega-3-polyunsaturated fatty acids reduces pathological retinal angiogenesis. Nat Med, 2007, 13, (7), 868-73. PMID:17589522.
- Lu, Y.*; Hong, S.*; Gotlinger, K.; Serhan, C. N., Lipid mediator informatics and proteomics in inflammation resolution. ScientificWorldJournal, 2006, 6, 589-614. PMID: 16752008. (*Share 1st authorship)
- Serhan, C. N.; Gotlinger, K.; Hong, S.; Lu, Y.; Siegelman, J.; Baer, T.; Yang, R.; Colgan, S. P.; Petasis, N. A., Anti-inflammatory actions of neuroprotectin D1/protectin D1 and its natural stereoisomers: assignments of dihydroxy-containing docosatrienes. J Immunol, 2006, 176, (3), 1848-59. PMID: 16424216.
- Duffield, J. S. *; Hong, S. *; Vaidya, V. S.; Lu, Y.; Fredman, G.; Serhan, C. N.; Bonventre, J. V., Resolvin D series and protectin D1 mitigate acute kidney injury.J Immunol, 2006, 177, (9), 5902-11, PMID: 17056514. PMID: 17056514. (*Share 1st authorship)
- Hong, S.; Tjonahen, E.; Morgan, E. L.; Lu, Y.; Serhan, C. N.; Rowley, A. F., Rainbow trout (Oncorhynchus mykiss) brain cells biosynthesize novel docosahexaenoic acid-derived resolvins and protectins-Mediator lipidomic analysis. Prostaglandins Other Lipid Mediat, 2005, 78, (1-4), 107-16. PMID: 16303609.
- Lu, Y.*; Hong, S.*; Tjonahen, E.; Serhan, C. N., Mediator-lipidomics: databases and search algorithms for PUFA-derived mediators. J Lipid Res, 2005, 46, (4), 790-802. PMID:15722568. (*Share 1st authorship)
- Arita, M.; Bianchini, F.; Aliberti, J.; Sher, A.; Chiang, N.; Hong, S.; Yang, R.; Petasis, N. A.; Serhan, C. N., Stereochemical assignment, antiinflammatory properties, and receptor for the omega-3 lipid mediator resolvin E1. J Exp Med, 2005, 201, (5), 713-22. PMCID:PMC2212834. PMID:15753205.
- Ariel, A.; Li, P. L.; Wang, W.; Tang, W. X.; Fredman, G.; Hong, S.; Gotlinger, K. H.; Serhan, C. N., The docosatriene protectin D1 is produced by TH2 skewing and promotes human T cell apoptosis via lipid raft clustering. J Biol Chem, 2005, 280, (52), 43079-86. PMID:16216871.
- Vance, R. E.; Hong, S.; Gronert, K.; Serhan, C. N.; Mekalanos, J. J., The opportunistic pathogen Pseudomonas aeruginosa carries a secretable arachidonate 15-lipoxygenase. Proc Natl Acad Sci U S A, 2004, 101, (7), 2135-9. 17. PMCID:PMC357064. PMID:14766977.
- Marcheselli, V. L.*; Hong, S.*; Lukiw, W. J.; Tian, X. H.; Gronert, K.; Musto, A.; Hardy, M.; Gimenez, J. M.; Chiang, N.; Serhan, C. N.; Bazan, N. G., Novel docosanoids inhibit brain ischemia-reperfusion-mediated leukocyte infiltration and pro-inflammatory gene expression. J Biol Chem, 2003, 278, (44), 43807-17. PMID:12923200. (*Share 1st authorship)
- Hong, S.; Gronert, K.; Devchand, P. R.; Moussignac, R. L.; Serhan, C. N., Novel docosatrienes and 17S-resolvins generated from docosahexaenoic acid in murine brain, human blood, and glial cells. Autacoids in anti-inflammation. J Biol Chem, 2003, 278, (17), 14677-87. PMID:12590139.
- Serhan, C. N.; Hong, S.; Gronert, K.; Colgan, S. P.; Devchand, P. R.; Mirick, G.; Moussignac, R. L., Resolvins: a family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. J Exp Med, 2002, 196, (8), 1025-37. PMCID:PMC2194036. PMID:12391014.
Additional Info
Protocols
Protocol for Hematoxylin and Eosin Staining
- The samples are sectioned into 20 um and stored in -20°C until using.
- Slides from each sectioned wound sample with the biggest wound gap are selected
- Immerse slides in acetone and store at -20C for 20 minutes.
- Remove slides from freezer and immerse in 100% ethanol for 2 minutes.
- Immerse in 95% ethanol for 2 minutes.
- Immerse in 80% ethanol for 2 minutes.
- Immerse in DI water for 2 minutes.
- Immerse in Mayer Hematoxylin for 30 seconds.
- Quickly remove from hematoxylin solution and rinse under tap until color no longer streaks across the slide.
- Immerse in DI water for 2 minutes.
- Dip quickly in 0.5% v/v acetic acid solution.
- Immerse in Eosin Y solution for 1 minute.
- Quickly remove from eosin solution and rinse under tap until color no longer streaks across the slide.
- Immerse in 95% ethanol for 2 minutes.
- Immerse in 100% ethanol for 2 minutes.
- Immerse in xylene for 5 minutes.
- Mount in a suitable xylene-based mounting media. Coverslip. Let air dry for 10 minutes.
- Seal the coverslip with clear nail polish. Air dry for 10 minutes.
- Store HE-stained slides at room temperature.
- Analyze using image J.
Protocol for Sirius Red Staining
- Immerse slides in acetone and store at -20C for 20 minutes.
- Make picro-sirius red solution (0.5 g Sirius red in 500 mL saturated picric acid).
- Remove slides from freezer and immerse in 100% ethanol for 2 minutes.
- Immerse in 95% ethanol for 2 minutes.
- Immerse in 80% ethanol for 2 minutes.
- Immerse in DI water for 2 minutes.
- Immerse in hematoxylin for 1 minute.
- Quickly remove from hematoxylin solution and rinse under tap until color no longer streaks across the slide.
- Immerse in DI water for 2 minutes.
- Immerse in picro-sirius red for one hour.
- Rinse under tap.
- Wash in two changes of acidified water (0.5% acetic acid v/v, 5mL in 1 L water).
- Immerse in DI water for 2 minutes.
- Immerse in 95% ethanol for 2 minutes.
- Immerse in 100% ethanol for 2 minutes.
- Immerse in xylene for 5 minutes.
- Mount in a suitable xylene-based mounting media. Coverslip. Let air dry for 10 minutes.
- Seal the coverslip with clear nail polish. Air dry for 10 minutes.
- Store Sirius-stained slides at room temperature in a labeled slide box with experiment info, name, date, etc.
Protocol for MASSON TRICHROME Staining
- Fix slides in 4% formalin – 30 mins
- Hydrate slides in DI water – wash 3X
- Preheat Bouin’s fluid in a water bath to 56-64°C in a fume hood.
- Place slides in the above solution for 60 mins followed by 10 min cooling.
- Rinse in tap water until sections are completely clear.
- Rinse once in DI water.
- Mix equal parts of Weigerts (A) and (B), and stain slides with working Weigert’s Iron Hematoxylin for 5-10 mins. Monitor and add stain as necessary to prevent drying.
- Rinse in running tap water for 2 mins.
- Rinse once in DI water.
- Apply Biebrich Scarlet/ Acid Fuchsin solution for 5-15 mins.
- Rinse in DI water.
- Differentiate in Phosphomolybdic/Phosphotungstic Acid solution for 10-15 mins or until collagen is not red.
- Without rinsing, apply Aniline Blue solution to the slides for 5-10 mins.
- Rinse slides in distilled water.
- Apply acetic acid solution (1%) to the slides for 3-5 mins.
- Dehydrate very quickly in 2 changes of 95% alcohol, followed by 2 chage of absolute alcohol.
- Clear in xylene, mount slides.
Procedures and Lipid Extraction Protocol from Cell Culture (MSC stem cells, control and treatment)
Method
- When MSC stem cells grow to 80% confluence, cells were used for lipid extraction.
- MSCs (three 10-cm dishes) were incubates in 50uM DHA in 4ml PBS for 20mins, and then treated with 10ng/ml TNF-alpha, 10ng/ml IL-1 and 100ng/ml LPS for 1hr. Control (three 10-cm dishes) was only incubates with 50uM DHA in PBS without stimulation.
- After incubation, 1ml ACN was added into solution and stored at -80°C
Lipid Extraction
- Use 2 volumes of ACN to stop the reaction in the cell culture in a 15-ml-tube for 1 ml culture. Adjust the pH to ~ 4.5-5.0 with ice-cold 0.1M HCl
- To each sample, add 50ul the internal standard containing 50 ng PGD2-d4
- Thaw the culture on ice.
- Sonicate the culture on ice for 10seconds/pulse x 5.
- Centrifuge at 5000rpm for 10min at 2°C
- Transfer the supernatant into a ice-cold 50ml-tube (tube 2)
- Add ~0.25ml ACN into tube 1 and sonicate cells
- Centrifuge at 5000rpm for 10min, transfer supernatant into tube 2
- Repeat 6,7 for twice.
- Add [pH5.0 NH4Ac-pH5 buffer] in tube 2 to bring the final concentration of ACN to 10%.
- Wash the SPE cartridge with 1 load of MeOH and then 1 load of deionized water
- Then pour the extraction in the cartridge and let it go through SPE slowly (dropwise).
Washing
- When liquid nearly runs out of SPE, turn off SPE, wash each cartridge with 3 loads [NH4Ac-pH5 buffer] to remove proteins.
- When liquid nearly runs out of SPE, turn off SPE, wash each cartridge with half load of [10%MeOH,NH4Ac-pH5 buffer] to remove yellow staff.
- Wash each cartridge with 0.5ml deionized-water(pH ~6)
- 1ml hexane was used to take away water.
- Let the air go through cartridge for 5min to dry water.
- Using Kimwipes to absorb the water attached at the tip of the SPE tubes and flow outlet.
Elute analytes out of SPE tubes
- Turn off the vacuum
- Put 10 ml-tip-attached-on-500ul-vials under the cartridges to collect ACN/methyl formate fractions.
- Add 1ml acetonitrile into SPE cartridge to elute lipids.
- Add 9ml 10% ACN/methyl formate (MF) to each cartridge.
- Let solvent soak into cartridge for about a minute before opening vacuum
- Open the vacuum gradually until 10% ACN/MF start to dripping slowly from the outlet of SPE tube
- Let 10% ACN/MF run through the SPE dropwise (i.e., slowly)
Nitrogen drying
- Use new glass tips
- Wash/rinse the 10ml tips with ~1-2ml of MeOH to remove the dust.
- Place the needle or glass tips ~1cm above the liquid surface of the sample.
- Dry down solvent gently till ~50 ul liquid left.
- Using 100 µl ACN/each wash to wash lipids on the wall of 10ml tip down into 500ul-vials
- Repeat 3 times: 1st, wash the top of the tip; 2nd, wash middle of tip; 3rd wash low end of tip into the 500ul-vials.
- Make sure no deposit in the solution. If seeing deposits, the solution may need to be filtered with 0.2um nylon syringe-filter.
- Dry down to final volume of 200ul.
- Vortex all vials to make sure that the analytes are dissolved and solutions are homogenous. Ready for LC-MS analysis
Protocol for Lipid Extraction from Tissues
- Sample tissues were cut into small 1-2 mm3 pieces. When cutting the samples, they were located on a plastic cap on ice.
- The small pieces were transferred to a 5mL tube.
- 1mL mixture of methanol and NH4Ac-pH4.5 (~10 mM) (MeOH-NH4AC-pH4.5=3:1) was added to each sample.
- pH was checked and changed to a range of 4.5. pH was regulated by adding 1mL of Acetic acid.
- 50µL of internal standard (PGD2-d4, NPD1-d2, and DHA-d5) with a concentration of 0.02ng each/µL were added to samples.
- Then the samples were homogenized using a probe sonicator. The tissues were homogenized for 1 minute [when homogenizing, the tubes were chilled by ice (inside the ice)].
- After homogenizing samples were centrifuged at 5000rpm for 15min at 4°C.
- Then the supernatant obtained was transferred into a new ice cold 50mL tube (Tube 2).
- 1mL of 100% methanol was added to the original tube, then samples were homogenized using the same procedure.
- Samples were centrifuged and supernatant transferred to the second tube.
- Steps 9 and 10 were repeated twice.
- The samples were left for 2 hours at -80°C in order to let any protein particles to precipitate.
- Then NH4Ac-pH4.5 buffer was added to tube 2, in order to bring the final methanol concentration to 10%.
SPE-Washing
- 500mg C18 SPE cartridges were conditioned.
- Add one load (9.45mL) of methanol; leave methanol for 10 min.
- Turn on the vacuum.
- Then let the methanol go through SPE, and add a load (9.45mL) of NH4AC-pH4.5 buffer. Let the NH4AC-pH4.5 buffer for 10 min in the cartridge.
- Then let the buffer go through the SPE and close it.
- The 10% Methanol/ NH4AC-pH4.5 buffer solution was poured to the cartridge and then the vacuum manifold was opened.
- The solution from above step 13 went through the SPE drop-wise.
- When the solution was about to be finished the SPE was closed and the cartridges washed with NH4AC-pH4.5 buffer. Cartridges were washed once with NH4AC-pH5.5 buffer solution.
- Then 1mL of 10% Methanol- NH4AC-pH4.5 buffer was added to each cartridge.
- Then 0.5mL deionized water (18 Ω) was poured to each cartridge.
- 1mL of Hexane was added to each cartridge, this in order to remove water and fat from the cartridge, if you need analyze DHA, avoid this step, just vacuum the cartridge for 5 min.
- Vacuum the cartridges for 5 min.
- The vacuum manifold tips were carefully dried with paper.
Elute Analytes out of SPE tubes
- Turn off the vacuum.
- Put 10ml coned glass tube under the cartridge. to collect MeOH fractions.
- Add 1ml MeOH to each cartridge.
- Add 9ml 20% MeOH/methyl formate (MF) to each cartridge.
- Let solvent soak in the cartridge for about a minute before opening vacuum.
- Open the vacuum gradually until 20% MeOH/MF (start to dripping slowly from the outlet of SPE tube
Nitrogen Drying
- Use new glass tips
- Place the needle or glass tips ~1cm above the liquid surface of the sample.
- Dry down solvent gently till ~50 ul liquid left.
- Using 100 µl METHANOL/each wash to wash lipids on the wall of 15ml-glass tube down to bottom, transfer methanol rinse into 500ul-vials
- Repeat 3 times: 1st, wash the top of the 15ml-glass-tube; 2nd, wash middle of middle of 15ml-glass-tube; 3rd wash low end of 15ml-glass-tube into the bottoms, then transfer to 500ul-vials.
- Make sure no deposit in the solution. If seeing deposits, the solution may need to be filtered with 0.2um nylon syringe-filter.
- Dry down to final volume of 50ul.
- Vortex all vials to make sure that the analytes are dissolved and solutions are homogenous. Ready for LC-MS analysis
- Label each vial with #, date, a brief description or abbreviations, and total ul.
Protocol for Lipid Extraction from Skin Wounds
- Samples were cut into small pieces. When cutting the samples, they were located on a plastic cap on ice.
- The small pieces were transferred to a 5mL tube.
- 1mL mixture of methanol and NH4AC-pH5.5 (MeOH-NH4AC-pH5.5=2:1) was added to each sample.
- pH was checked and changed to a range of 4.5-5.5. pH was regulated by adding 1mL of Acetic acid.
- 50µL of internal standard (PGD2-d4) with a concentration of 0.1ng/µL were added to samples.
- Then the samples were homogenized using a probe sonicator. The tissues were homogenized for 1 minute (when homogenizing, the tubes were close to the ice).
- After homogenizing samples were centrifuged at 5000rpm for 15min at 4°C.
- Then the supernatant obtained was transferred into a new ice cold 50mL tube (Tube 2).
- 1mL of 100% ACETONE was added to the original tube, then samples were homogenized using the same procedure before discussed.
- Samples were centrifuged and supernatant transferred to the second tube.
- Steps 9 and 10 were repeated twice.
- The samples were left overnight at -80°C; this in order to let any protein particles to precipitate.
- Then NH4AC-pH5.5 buffer was added to tube 2, in order to bring the final acetone concentration to 10%.
SPE-Washing
- 500mg cartridges were reconditioned.
- Add one load (9.45mL) of methanol; leave methanol for 10 min.
- Turn on the vacuum.
- Then let the methanol go through SPE, and add a load (9.45mL) of NH4AC-pH5.5 buffer. Let the NH4AC-pH5.5 buffer for 10 min in the cartridge.
- Then let the buffer go through the SPE and close it.
- The 10% Methanol/ NH4AC-pH5.5 buffer solution was poured to the cartridge and then the vacuum manifold was opened.
- The solution went through the SPE drop wise.
- When the solution was about to be finished. The SPE was closed and the cartridges washed with NH4AC-pH5.5 buffer. Cartridges were washed once with NH4AC-pH5.5 buffer solution.
- Then 1mL of 10% Methanol- NH4AC-pH5.5 buffer was added to each cartridge.
- Then 0.5mL deionized water (18 Ω) was poured to each cartridge.
- 1mL of Hexane was added to each cartridge, this in order to remove water from the cartridge.
- Air passed through cartridges for 5 min.
- The vacuum manifold tips were carefully dried with paper.
- Cartridges were used 3 times before being disposed.
Elute Analytes out of SPE tubes
- The SPE manifold top was moved to another glass manifold.
- 10mL tips were attached to 250µL vials, under the cartridge.
- 1mL of methanol was poured to each cartridge, to elute lipids.
- 9mL of 10% MeOH/methyl formate were used to wash tubes and then this solution was poured to the cartridges.
- The solution soaked into the cartridge for 1min before opening the vacuum.
- Gradually, the vacuum was opened until the solution started dripping out; it must be done in a drop wise manner.
- The solution ran through the SPE.
Nitrogen Drying
- New glass tips were attached to the nitrogen dryer.
- Nitrogen went through the tips in order to remove dust.
- The tips were place at a level where nitrogen could blow the solution surface (about 1cm above the liquid surface).
- Solvent was dried until it had a total volume of 100µL.
- 200µL methanol was used to wash lipids from the 10mL tip’s wall.
- The top of the tip was first washed
- Then the middle area
- At the end the lower tip area
- The solution was dried to a final volume of 100µL.
- The vials were vortexed; this to make sure that the analytes were dissolved and the solutions were homogenous.
- 200 µL of H2O were added to the total volume.
- Samples are kept in a storing box at -80°C.
Protocol for Lipid Extraction from db/db Diabetic and db+ Non-diabetic Macrophages
Incubation:
- Prepare macrophages from mice following the published protocols.
- Place the cells collected from mice in culture dishes.
- Washed away non-attached cells.
- Take pictures for attached cells, 3 fields were chosen for each dish
- Incubated macrophages with 5ug DHA in 3ml PBS for each dish for 20min.
- Then stimulate cells with 10ng/ml TNF-alpha, IL-1 beta, and 100ng/ml LPS for 1h.
- Add 1ml ACN into dish to stop reaction and stored at -80 degree.
- Calculate the cell number/field as cell ratio.
Lipid Extraction
- To each sample, add 50 ng PGD2-d4 (1ng/ul)
- Thaw the culture on ice.
- Centrifuge at 5000rpm for 10min at 4°C
- Transfer the supernatant into a ice-cold 50ml-tube (tube 2)
- Add 0.25ml ACN into tube 1 and sonicate cells
- Sonicate the culture on ice for 10seconds/pulse x 5.
- Centrifuge at 5000rpm for 10min, transfer supernatant into tube 2
- Repeat 6,7 for twice.
- Add [NH4Ac-pH5 buffer] in tube 2 to bring the final concentration of ACN to 10%.
- Go through SPE slowly (dropwise).
Washing
- When liquid nearly runs out of SPE, turn off SPE, wash each cartridge with 3 loads [NH4Ac-pH5 buffer] to remove proteins.
- When liquid nearly runs out of SPE, turn off SPE, wash each cartridge with half load of [10%MeOH,NH4Ac-pH5 buffer] to remove yellow staff.
- Wash each cartridge with 0.5ml deionized-water(pH ~6)
- 1ml hexane was used to take away water.
- Let the air go through cartridge for 5min to dry water.
- Using Kimwipes to sorb the water attached at the tip of the SPE tubes and flow outlet
Elute analytes out of SPE tubes
- Turn off the vacuum
- Put 10 ml TIP-ATTACHED-ON-500ul-VIALS under the cartridges to collect ACN/methyl formate fractions.
- Add 1ml acetonitrile into SPE cartridge to elute lipids.
- Add 9ml 10% ACN/methyl formate (MF) to each cartridge.
- let solvent soak into cartridge for about a minute before opening vacuum
- Open the vacuum gradually until 10% ACN/MF (start to dripping slowly from the outlet of SPE tube
- Let 10% ACN/MF run through the SPE dropwise (i.e., slowly)
Nitrogen drying
- Use new glass tips
- Wash/rinse the 10ml tips with ~1-2ml of MeOH to remove the dust.
- Place the needle or glass tips ~1cm above the liquid surface of the sample.
- Dry down solvent gently till ~50 ul liquid left.
- Using 100 µl ACN/each wash to wash lipids on the wall of 10ml tip down into 500ul-vials
- Repeat 3 times: 1st, wash the top of the tip; 2nd, wash middle of tip; 3rd wash low end of tip into the 500ul-vials.
- Make sure no deposit in the solution. If seeing deposits, the solution may need to be filtered with 0.2um nylon syringe-filter.
- Dry down to final volume of 200ul.
- Vortex all vials to make sure that the analytes are dissolved and solutions are homogenous. Ready for LC-MS analysis
- Label each vial with #, date, a brief description or abbreviations, and total ul.
Mass Spectrometry Data for Lipid Mediators and Hodrogels
- Hong S*, Baravkar SB, Lu Y, Masoud AR, Zhao Q, Zhou W. Molecular Modification of Queen Bee Acid and 10-Hydroxydecanoic Acid with Specific Tripeptides: Rational Design, Organic Synthesis, and Assessment for Prohealing and Antimicrobial Hydrogel Properties. Molecules. 2025;30(3). doi: 10.3390/molecules30030615. PubMed PMID: 39942719; PubMed Central PMCID: PMC 11819776.
- Lu Y, Su S, Chu CC, Kobayashi Y, Masoud AR, Peng H, Lien N, He M, Vuong C, Tran R, Hong S*. Amino Acid-Based Protein-Mimic Hydrogel Incorporating Pro-Regenerative Lipid Mediator and Microvascular Fragments Promotes the Healing of Deep Burn Wounds. International journal of molecular sciences. 2024;25(19). doi: 10.3390/ijms251910378. PubMed PMID: 39408708; PubMed Central PMCID: PMC11476471.
- Lu Y, Tian H, Peng H, Wang Q, Bunnell BA, Bazan NG, Hong S*. Novel lipid mediator 7S,14R-docosahexaenoic acid: biogenesis and harnessing mesenchymal stem cells to ameliorate diabetic mellitus and retinal pericyte loss. Frontiers in cell and developmental biology. 2024;12:1380059. doi: 10.3389/fcell.2024.1380059. PubMed PMID: 38533089; PubMed Central PMCID: PMC10963555.
- Baravkar SB, Lu Y, Masoud AR, Zhao Q, He J, Hong S*. Development of a Novel Covalently Bonded Conjugate of Caprylic Acid Tripeptide (Isoleucine-Leucine-Aspartic Acid) for Wound-Compatible and Injectable Hydrogel to Accelerate Healing. Biomolecules. 2024;14(1). doi: 10.3390/biom14010094. PubMed PMID: 38254694; PubMed Central PMCID: PMC10813153.
- Ogawa N, Sone S, Hong S, Lu Y, Kobayashi Y. Synthesis of Two Stereoisomers of Potentially Bioactive 13,19,20-Trihydroxy Derivative of Docosahexaenoic Acid. Synlett : accounts and rapid communications in synthetic organic chemistry 2020, 31:1735-9. PMCID: PMC8752060, PMID:35023886
- Hong S*, Lu Y, Morita M, Saito S, Kobayashi Y, Jun B, Bazan NG, Xu X, Wang Y. Stereoselective Synthesis of Maresin-like Lipid Mediators. Synlett : accounts and rapid communications in synthetic organic chemistry 2019, 30:343-7. PMID:31086432.
- Nishimura K, Sakaguchi T, Nanba Y, Suganuma Y, Morita M, Hong S, Lu Y, Jun B, Bazan NG, Arita M, Kobayashi Y. Stereoselective Total Synthesis of Macrophage-Produced Prohealing 14,21-Dihydroxy Docosahexaenoic Acids. The Journal of organic chemistry 2018, 83:154-66. PMID:29224348.
- Bazan HA, Lu Y, Jun B, Fang Z, Woods TC, Hong S. Circulating inflammation-resolving lipid mediators RvD1 and DHA are decreased in patients with acutely symptomatic carotid disease. Prostaglandins, leukotrienes, and essential fatty acids 2017, 125:43-7. PMCID: PMC5909403 PMID:28987721.
- Hong S*, Tian H, Lu Y, Laborde JM, Muhale FA, Wang Q, Alapure BV, Serhan CN, Bazan NG. Neuroprotectin/protectin D1: endogenous biosynthesis and actions on diabetic macrophages in promoting wound healing and innervation impaired by diabetes. American journal of physiology Cell physiology 2014, 307:C1058-67. PMCID: PMC4254953, PMID:25273880.
- Hong S*, Lu Y, Tian H, Alapure BV, Wang Q, Bunnell BA, Laborde JM. Maresin-like lipid mediators are produced by leukocytes and platelets and rescue reparative function of diabetes-impaired macrophages. Chemistry & biology 2014, 21:1318-29. PMCID: PMC4224612 PMID:25200603.
- Tian H, Lu Y, Shah SP, Hong S*. Autacoid 14S,21R-dihydroxy-docosahexaenoic acid counteracts diabetic impairment of macrophage prohealing functions. The American journal of pathology 2011, 179:1780-91. PMCID:PMC3181353, PMID:21839062
- Tian H, Lu Y, Shah SP, Hong S*. 14S,21R-Dihydroxydocosahexaenoic Acid Remedies Impaired Healing and Mesenchymal Stem Cell Functions in Diabetic Wounds. The Journal of biological chemistry 2011, 286:4443-53. PMCID:PMC3039401, PMID:21112969.
- Lu Y, Tian H, Hong S*. Novel 14,21-dihydroxy-docosahexaenoic acids: structures, formation pathways, and enhancement of wound healing. Journal of lipid research 2010, 51:923-32. PMCID:PMC2853460, PMID:19965612.

 myLSUHSC
myLSUHSC